How to use Glycan Fragment DB
Here we describe how to search Glycan Fragment DB (GFDB) and what the search result means in a step-by-step manner. For illustration purposes, we will use the following glycan sequence as an example.
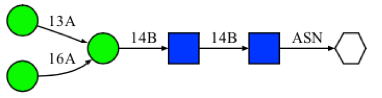
How to search Glycan Fragment DB
- The search box will begin with following form.
-
Use the "+" buttons to add and select an appropriate carbohydrate sequence. Following image illustrates how to build the example glycan sequence in a stepwise manner. As a glycan sequence is being modified by the user, the glycan sequence diagram will be updated accordingly to provide visual guidance to the user. To construct the branched sequence, press the "+" button at the branching point.
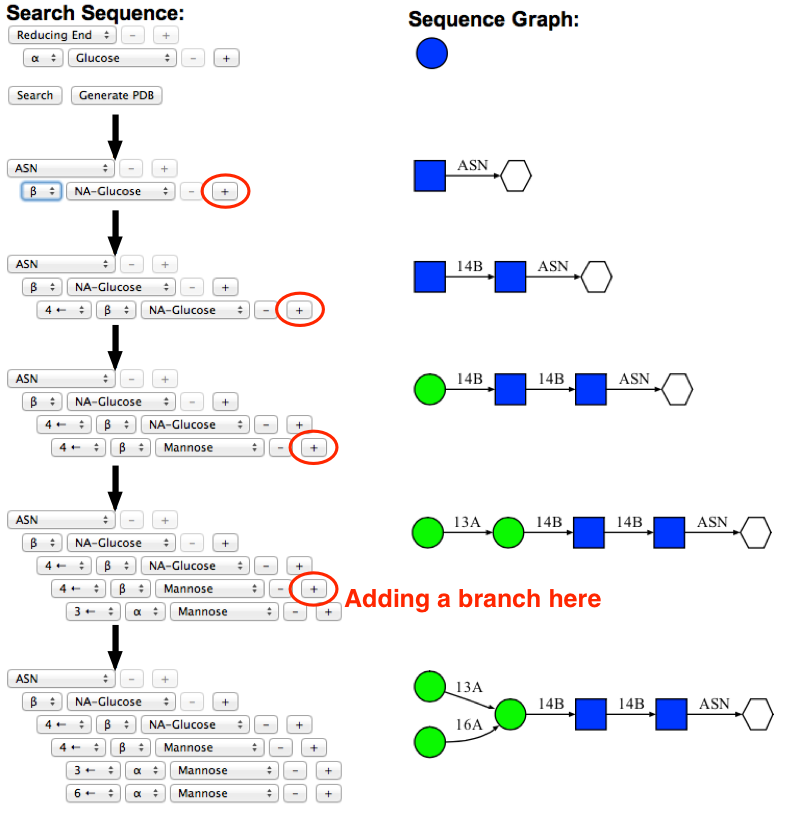
-
Once the desired glycan sequence is built in the form, press "Search" button to perform database search. When the search is completed, the torsion angle distribution for the first glycosidic linkage is generated and displayed. The selected glycosidic linkage will be highlighted in red.
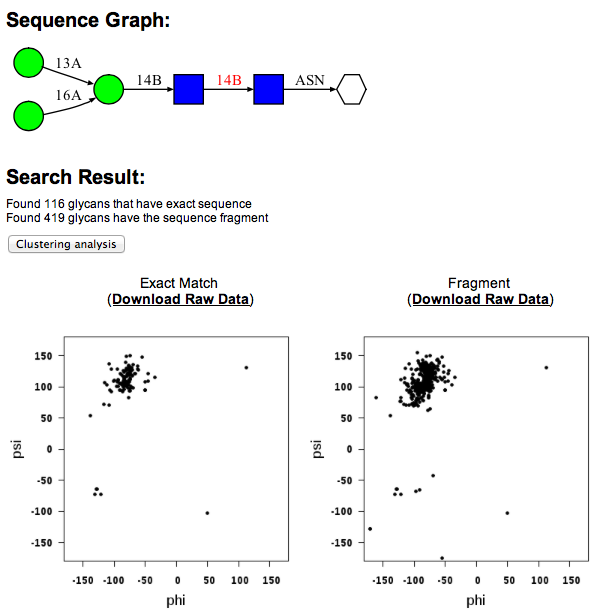
The search result indicates that there are 119 suctures that match exactly with the example sequence. In addition, there are 419 glycan fragment structures that match with search sequence. A "fragment" structure is a glycan substructure that matches with the given sequence but is a part of larger glycan structure. For example, the highlighted portions of the following glycan structures are glycan "fragment" structures that match with the example sequence.
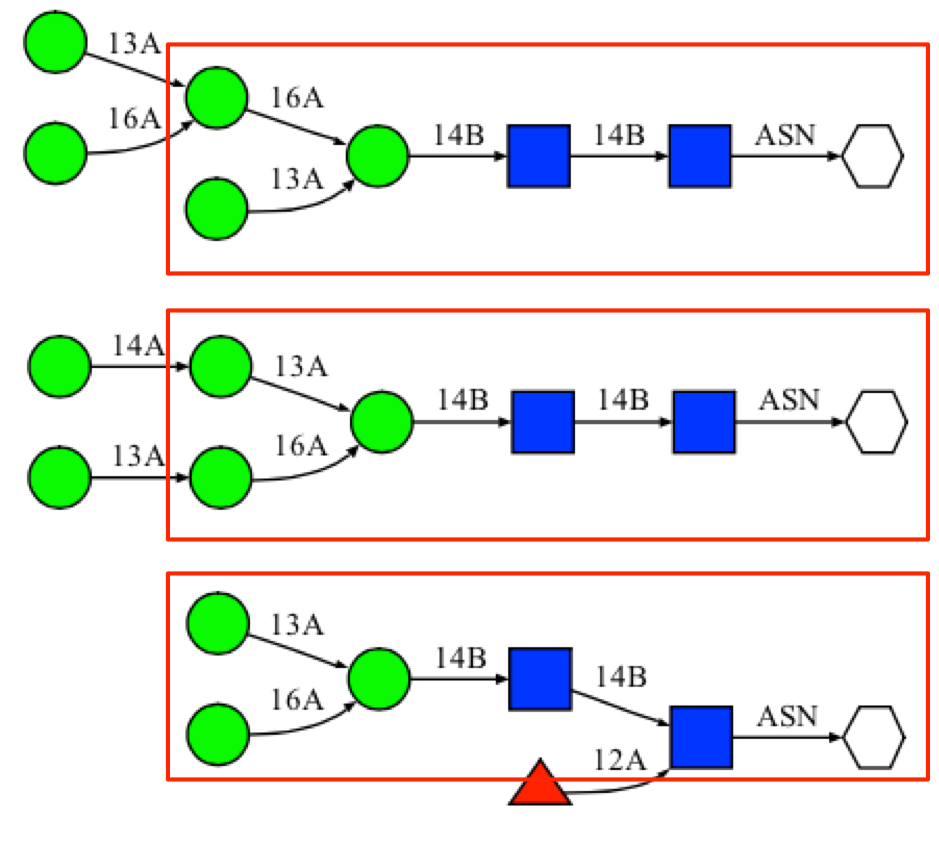
-
Torsion angle results for different glycosidic linkages can be displayed by clicking the glycosidic linkages in the sequence diagram (click the highligted area in the following image). Raw data can be downloaded in a text format file using the "Raw Data" link above the torsion angle distribution map.
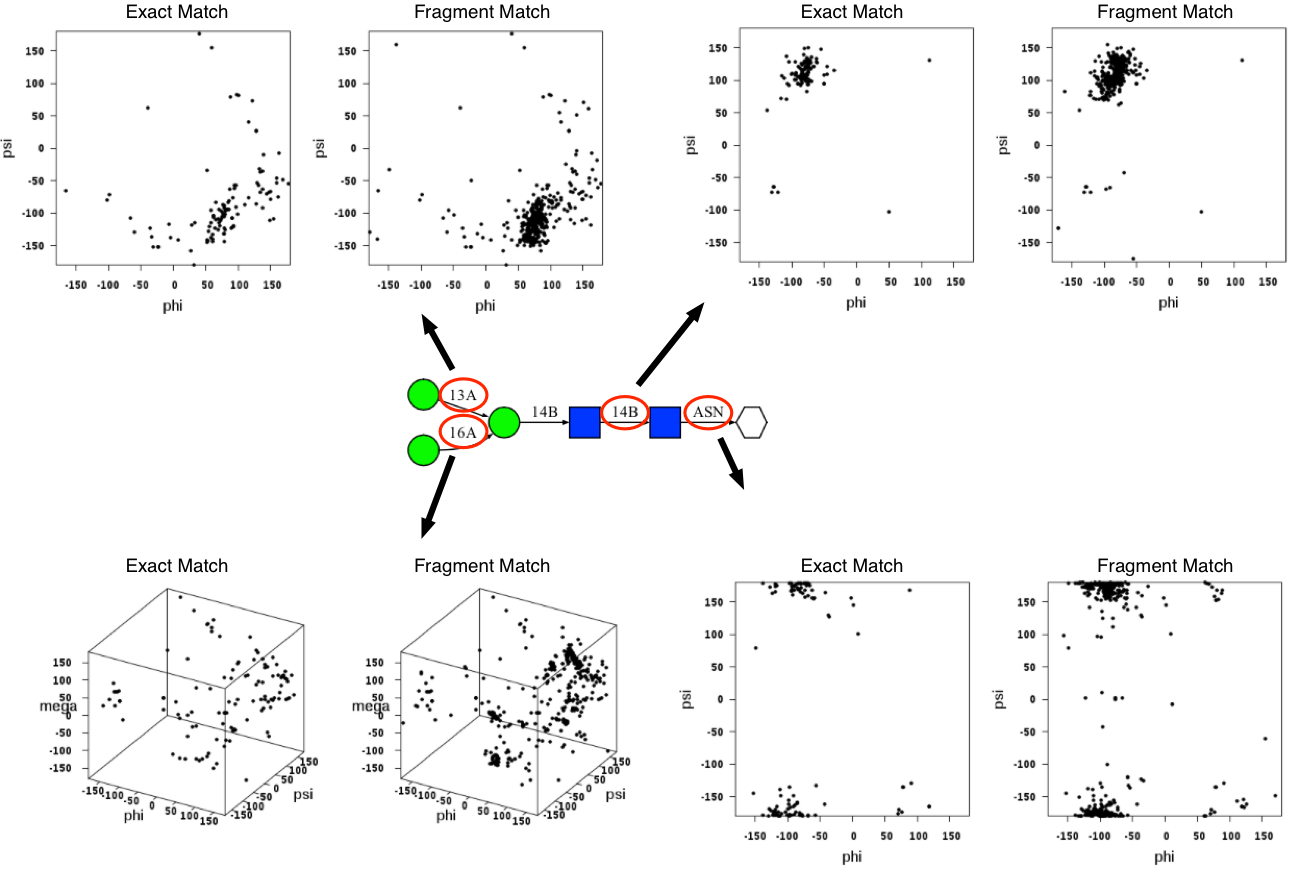
The glycosidic torsion angle definition is adopted from the crystallographic definition; O5-C1-O1-C'x (Φ; phi), C1-O1-C'x-C'x-1 (Ψ; psi), and O1-C'6-C'5-O'5 (ω; omega). The torsion angle between the first residue of the N-glycan chain and the side chain of the asparagine residue is defined as O5-C1-N'D2-C'G (Φ; phi) and C1-N'D2-C'G-C'B (Ψ; psi). The torsion angle between the first residue of the O-glycan chain and the side chain of the serine residue is defined as O5-C1-O'G-C'B (Φ; phi) and C1-O'G-C'B-C'A (Ψ; psi). For threonine residue, OG1 is used instead of OG. The atom names are based on the CHARMM topology.
Filter options
GFDB provides various filters for a glycan structure search.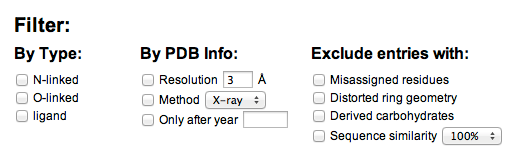
- Filter by glycosidic type: GFDB will only returns the glycosidic torsion angle results from those glycans that are matched with selected types. For example, when the N-linked type filter is selected, the results from glycans that are either O-linked or ligand will be excluded.
- Filter by PDB infomation: GFDB will only return the glycosidic torsion angle results from the glycans in the PDB that are matched with selected filters. For example, when the 3Å resolution filter is selected, the result will only be consists of glycan structures whose X-ray resolutions are smaller than 3Å.
- Excludes entries: Unlike the other filter options, this option will excludes entries that match with the selected options.
- "Excludes emisassigned residue" will removes the entries that have mismatch between the residue name annotation in the PDB file and the chemical name determined by the actual coordinates of the molecule. GFDB stores the residue names derived by the actual coordinates, which is more important information thant the residue name annotation in the PDB, and such mismatch could be a simple mistake in the PDB annotation. Thus, not all entries having mismatch are not errerneous cases of glycan structures themselves. Nonetheless, such residue name mismatch could be a potential error, and the entries contain residue name mismatch will be excluded with this option.
- "Excludes distorted geometry" will removes the entries that have carbohydrate monomers having conformation other than chair conformation (1C4 or 4C1). Distortions in the carbohydrate ring geometry can interfere with the torsion angle analysis results, and the entries that contain residues with distorted geometry will be excluded.
- "Excludes derived carbohydrate" will removes the entries that have chemically modified carbohydrates (e.g. phosphorylation, methylation, etc)
- "Excludes sequence similarity" will use protein sequence similarity database provided by RCSB (for more informations, click here) and excludes entries having sequence similarity higher than the selected option. Due to difficulties in determining the protein that is associated with ligand-type glycans, this option is only applied to N-linked or O-linked glycans.
Clustering analysis
-
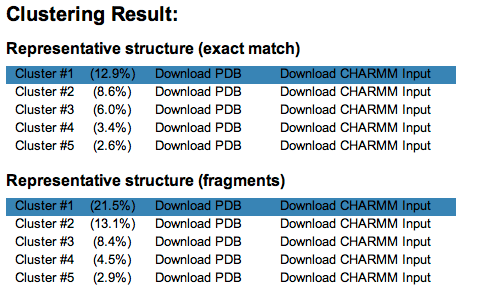
Clustering analysis of resulting glycosidic torsion angles can be performed by clicking the "Clustering analysis" button above the torsion angle distribution plots. The root-mean-square difference of glycosidic torsion angles are used as a distance metric in the clustering analysis and a cutoff of 30 (similar to 30 degree) is used as a default. A simple clustering method is applied; the first cluster is selected as a glycan entry having the largest number of neighbors and its neighbors are selected as the first cluster, and the seond cluster is selected in the same manner after the members of the first cluster is excluded. Once the clusters are determined, the average torsion angle values are calculated among the cluster members and the corresponding 3D structure of the glycans are generated. Use the "download PDB" link to download the representative structures of cluster. Selected clusters will be highlighted.
Generating a report
-
Because searching and clustering analysis may take a long time (up to 5 minutes), we provide a way to generate an archive file that contains all the raw data (glycosidic torsion angles, figures, and clustering analysis result). You can either download the archived file after a report is generated or receive an e-mail containing the report (if e-mail address is provided). E-mail will be sent even if the browser is closed or the user has left the page.
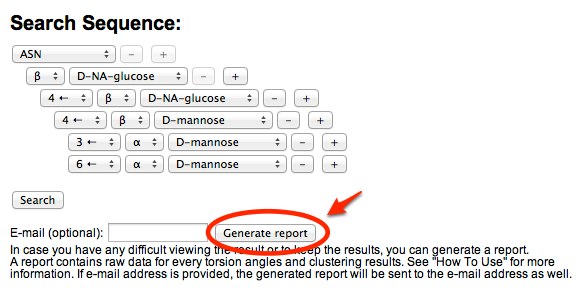
-
The archived file contains several files, and some of the important files are discussed below. An example of the generated report can be downloaded here.
- sequence.txt: This file contains the glycan sequence being searched in a text format. When there are more than one glycosidic linkages, each linkage is numbered as they appeared in the sequence file starting from 1. (in the case of N-liked or O-linked glycan, a linkage number of 0 is assigned to the linkage between the protein and the first carbohydrate residue).
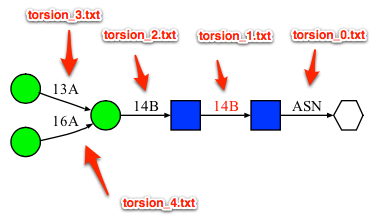

- sequence/ or fragment/ directory: sequence/ folder contains the results based on the exact match and fragment/ folder contains the results from the fragment match.
- sequence/torsion_X.txt or fragment/torsion_X.txt: This file contains list of glycosidic torsion angles for the exact or fragment matches, respectively. X in the file name refers to the glycosidic linkage number. See above image for an example.
- sequence/clust_X.pdb or fragment/clust_X.pdb: This file contains the 3D structure of glycans based on the clustering analysis result. X in the file name refers to different clusters (X = 1 for the largest cluster).
- sequence/clust_X_torsion_Y.txt or fragment/clust_X_torsion_Y.txt: This file contains the list of torsion angles that belong to a cluster. X in the filename refers to the cluster number and Y in the filename refers to the glycosidic linkage number.
- sequence.txt: This file contains the glycan sequence being searched in a text format. When there are more than one glycosidic linkages, each linkage is numbered as they appeared in the sequence file starting from 1. (in the case of N-liked or O-linked glycan, a linkage number of 0 is assigned to the linkage between the protein and the first carbohydrate residue).
