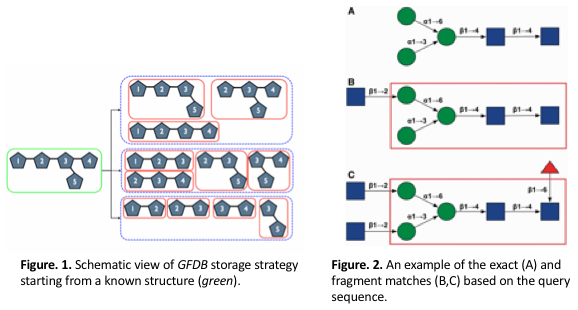
About Glycan Fragment DB
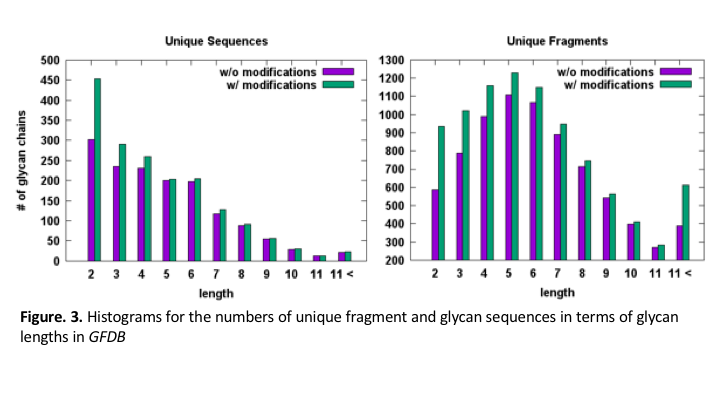
We have developed Glycan Fragment DB (GFDB) by searching the entire PDB, identifying PDB structures with biologically relevant carbohydrate moieties, and classifying PDB glycan structures based on their primary sequence and glycosidic linkage. After a glycan search is complete, each glycosidic torsion angle distribution is displayed in terms of the exact match and the fragment match. The exact match results are from the PDB entries that contain the glycan sequence identical to the query sequence. The fragment match results are from the entries with the glycan sequence whose substructure (fragment) or entire sequence is matched to the query sequence, such that the fragment results implicitly include the influences from the nearby carbohydrate residues (Figure 2). Figure 1 illustrates a schematic view of hierarchical building procedure that we have developed for the GFDB; e.g., starting from a known glycan with sugar 1-2-(3-5)-4 (green box) to fragments enclosed by each red box. As of December 2017, for the glycan structures with more than 2 carbohydrates, the hierarchical fragmentation identified a total of 127,202 fragment structures with 1,754 unique glycan sequences in the PDB; a unique glycan sequence has more than 2 carbohydrates and is defined by the carbohydrate sequence and the glycosidic linkages. There are 1,754 unique sequences and 9,055 unique fragments with chemical modifications. Without considering chemical modifications, the number of unique sequences and fragments are 1,492 and 7,741 (Figure 3).